We develop cutting edge technologies – many of which utilize sequencing-based technology – to both map and perturb chromatin state, with a focus on single cells, single molecules, and large scale perturbations with small molecules and CRISPR screens. Our current projects focus on:
- High throughput low input and single cell transcriptomic and epigenomic mapping
- Single cell and single molecule direct readout of RNA and chromatin modifications
- Combinatorial CRISPR screens
- Small molecule screens
People
- Private: Sofia Battaglia, PhD
- Volker Hovestadt, PhD
- Sarah Shareef
- Chuck Epstein, PhD
- Noam Shoresh, PhD
- Kevin Grosselin, PhD
Publications
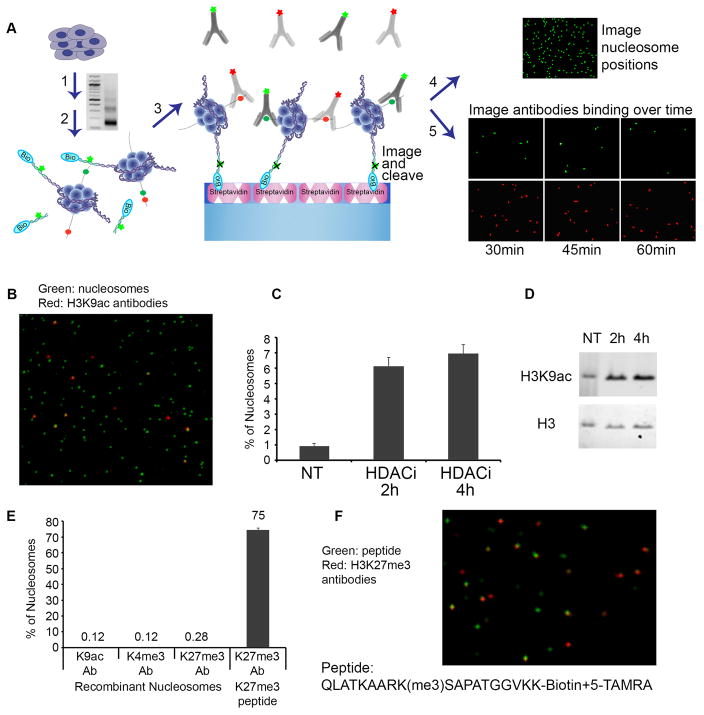
Want to know more about this aspect of the lab? We recommend starting with the papers belowDifferent combinations of histone modifications have been proposed to signal distinct gene regulatory functions, but this area is poorly addressed by existing technologies. We applied high-throughput single-molecule imaging to decode combinatorial modifications on millions of individual nucleosomes from pluripotent stem cells and lineage-committed cells. We identified definitively bivalent nucleosomes with concomitant repressive and activating marks, … Continued
Combinatorial genetic screening using CRISPR-Cas9 is a useful approach to uncover redundant genes and to explore complex gene networks. However, current methods suffer from interference between the single-guide RNAs (sgRNAs) and from limited gene targeting activity. To increase the efficiency of combinatorial screening, we employ orthogonal Cas9 enzymes from Staphylococcus aureus and Streptococcus pyogenes. We … Continued
Genome-wide profiling of histone modifications can provide systematic insight into the regulatory elements and programs engaged in a given cell type. However, conventional chromatin immunoprecipitation and sequencing (ChIP-seq) does not capture quantitative information on histone modification levels, requires large amounts of starting material, and involves tedious processing of each individual sample. Here, we address these … Continued
Chromatin profiling provides a versatile means to investigate functional genomic elements and their regulation. However, current methods yield ensemble profiles that are insensitive to cell-to-cell variation. Here we combine microfluidics, DNA barcoding and sequencing to collect chromatin data at single-cell resolution. We demonstrate the utility of the technology by assaying thousands of individual cells and … Continued
